About me
Contact : madrid AT sat.t.u-tokyo.ac DOT jp
Since April 2024 I am postdoc in the team of Tetsuya Kobayashi at the Institute of Industrial Science of The University of Tokyo.
Until February 2024 I was a PhD student in the group PEIPS (Population Evolution and Interacting Particle Systems) and Inria project team MERGE of CMAP (École polytechnique, CNRS UMR 7641), under the supervision of Sylvie Méléard and Meriem El Karoui. Until March 2024 I continued my research founded by the ERC project SINGER.
Research topics
I’m a probabilist applied mathematician motivated by the interplay between the different scales of living systems, from the dynamics of chemical reaction networks to the evolution of populations. I am particularly interested by questions concerning the long time behaviour of those systems (stability, balanced growth, large deviations).
Some keywords:
- Non-local branching processes
- Stochastic Individual-Based Models
- Structured Population Dynamics
- Single-cell modelling and inference in microbiology
Ongoing projects
Populations with reinforced phenotypic plasticity
Biological systems choose actions without an explicit reward signal (intrisic motivation). Recent works apply this idea to design robust optimisation algorithms that efficiently explore the state space rather than selecting a unique optimal solution. Our goal is to generalise the framework of ancestral learning to propose and study selection-mutation algorithms that exhibit intrinsic motivation.
Questions and current work:
- Characterise the trajectories of a population that combines ancestral reinforcement and ancestral curiosity:
- Describe its long-time behaviour (quasi-stationary distributions, typical trajectories (spinal decomposition)).
- Using the pathwise representation, define and solve optimal control problems that select the “most curious” trajectory (ex. maximise entropy, maximise space coverage)
Large deviations of growing stochastic systems of chemical reactions
With Praful Gagrani, on the large deviations of functionals of some chemical reaction networks, with applications to growing cells.
Emergence and stability of cell growth under stress: from single cells to populations
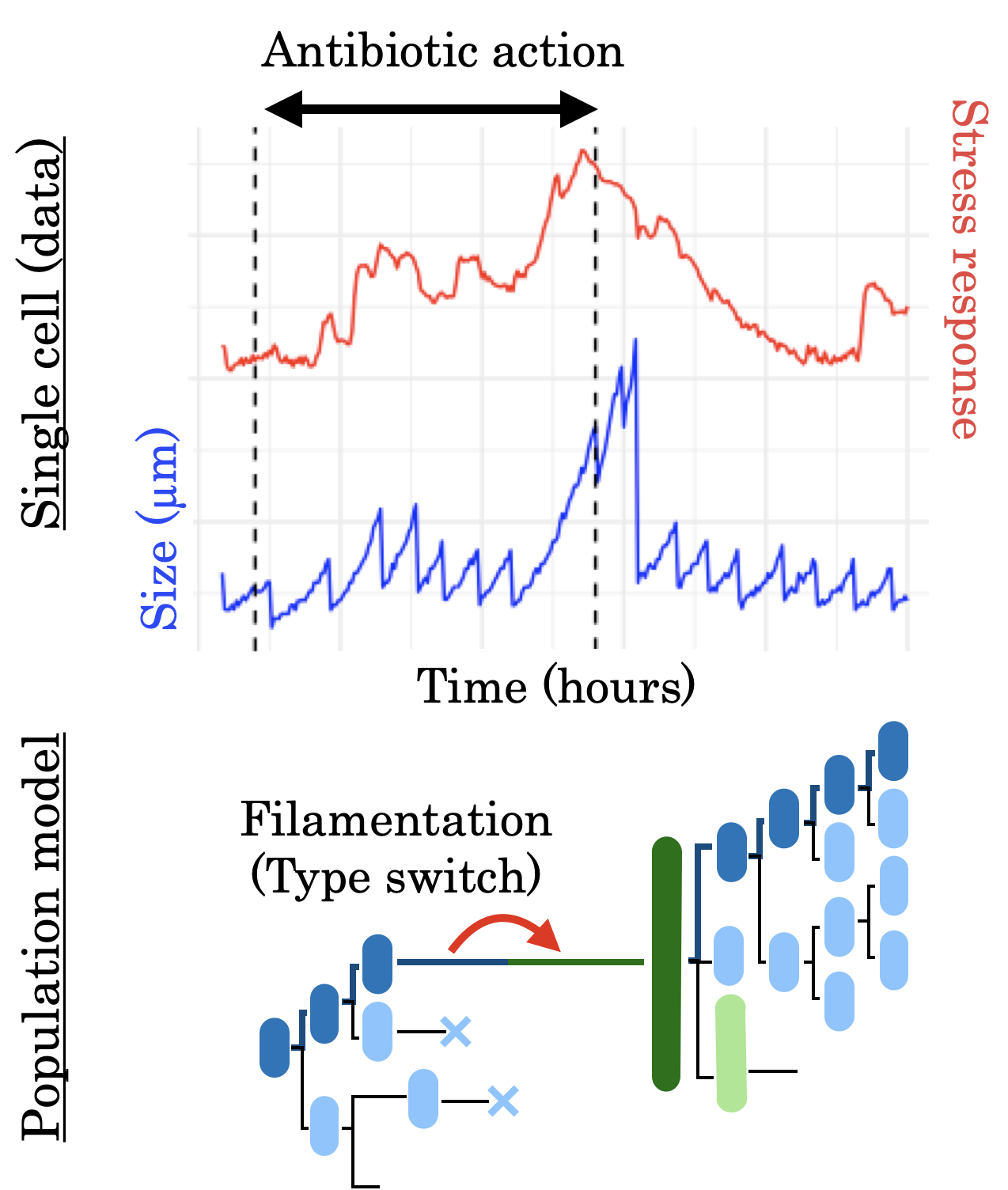
In my PhD, I aimed to model the growth and emergence of heterogeneity in E. coli bacterial populations under antibiotic stress. You can found my thesis here.
Under the action of an antibiotic damaging DNA, bacteria change their growth strategy and do not respond homogeneously to the stress. In addition, the effect of the antibiotic is different depending on the speed of bacterial growth which itself depends on the richness of the growth medium. Using measure-valued stochastic processes to model structured population dynamics I studied the emergence of bacterial subpopulations exhibiting different behaviors depending on genotoxic stress and on the nutritional richness of the environment in which they are immersed.
Single electron counting statistics for DNA-based biosensors
In collaboration with Nicolas Clément (LIMMS CNRS - UTokyo), we are interested in the estimation of the probability distribution associated to the current of single-electron electrical measures in DNA-based biosensors developped in Prof. Fujii’s lab.
.JPG)



